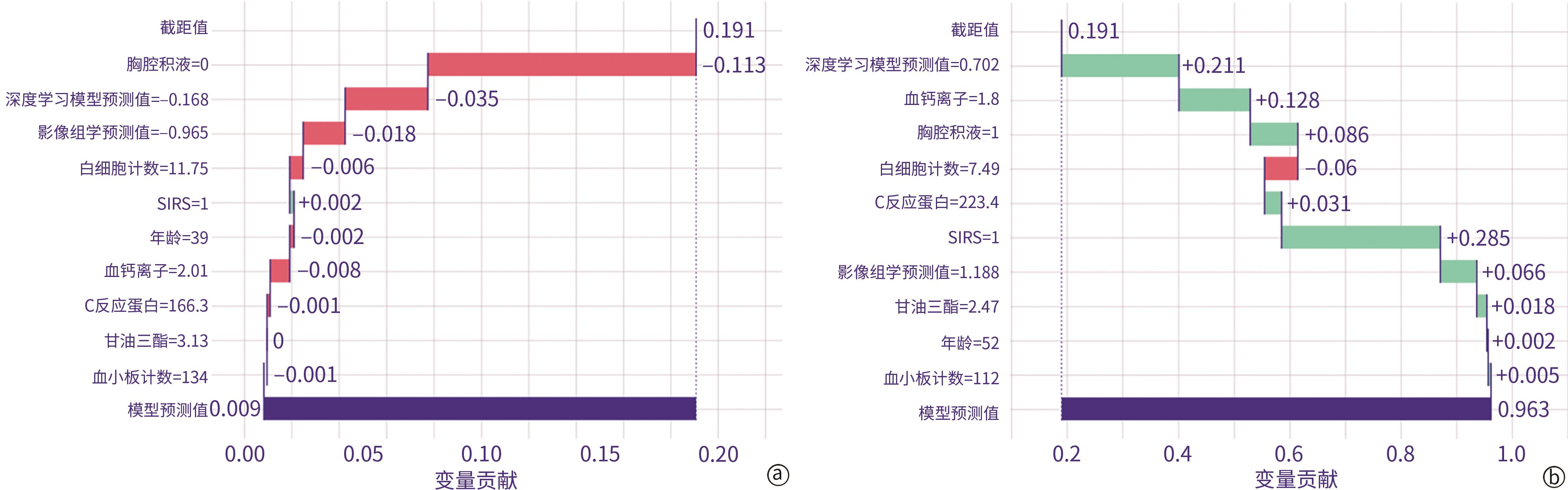
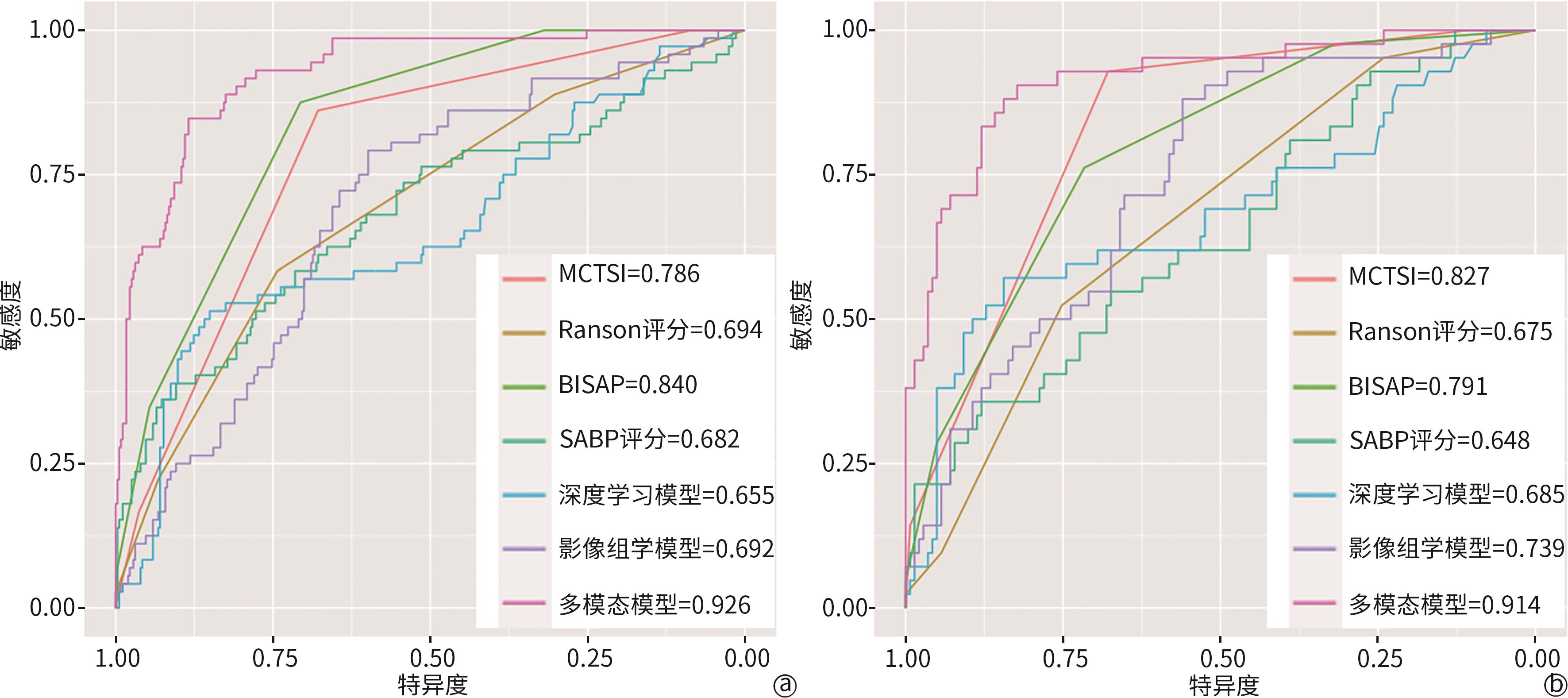
| [1] |
PETROV MS, YADAV D. Global epidemiology and holistic prevention of pancreatitis[J]. Nat Rev Gastroenterol Hepatol, 2019, 16( 3): 175- 184. DOI: 10.1038/s41575-018-0087-5. |
| [2] |
TRIKUDANATHAN G, YAZICI C, EVANS PHILLIPS A, et al. Diagnosis and management of acute pancreatitis[J]. Gastroenterology, 2024, 167( 4): 673- 688. DOI: 10.1053/j.gastro.2024.02.052. |
| [3] |
|
| [4] |
YIN MY, ZHU JD, LIU L, et al. Research advances in machine learning models for acute pancreatitis[J]. J Clin Hepatol, 2023, 39( 12): 2978- 2984. DOI: 10.3969/j.issn.1001-5256.2023.12.034. |
| [5] |
XIE F, ZHANG KQ, LI F, et al. Diagnostic accuracy of convolutional neural network-based endoscopic image analysis in diagnosing gastric cancer and predicting its invasion depth: A systematic review and meta-analysis[J]. Gastrointest Endosc, 2022, 95( 4): 599- 609. DOI: 10.1016/j.gie.2021.12.021. |
| [6] |
HAGGENMÜLLER S, MARON RC, HEKLER A, et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts[J]. Eur J Cancer, 2021, 156: 202- 216. DOI: 10.1016/j.ejca.2021.06.049. |
| [7] |
LI GW, LIU J, CAO H, et al. Research review of deep learning in colon polyp image segmentation[J]. J Front Comput Sci Technoly, 2025, 19( 5): 1198- 1216. DOI: 10.3778/j.issn.1673-9418.2408012. |
| [8] |
PACAL I, KARABOGA D. A robust real-time deep learning based automatic polyp detection system[J]. Comput Biol Med, 2021, 134: 104519. DOI: 10.1016/j.compbiomed.2021.104519. |
| [9] |
YU T, LIN N, ZHANG X, et al. An end-to-end tracking method for polyp detectors in colonoscopy videos[J]. Artif Intell Med, 2022, 131: 102363. DOI: 10.1016/j.artmed.2022.102363. |
| [10] |
LIU H, ZHUANG YZ, SONG EM, et al. A 3D boundary-guided hybrid network with convolutions and Transformers for lung tumor segmentation in CT images[J]. Comput Biol Med, 2024, 180: 109009. DOI: 10.1016/j.compbiomed.2024.109009. |
| [11] |
BANKS PA, BOLLEN TL, DERVENIS C, et al. Classification of acute pancreatitis: 2012: Revision of the Atlanta classification and definitions by international consensus[J]. Gut, 2013, 62( 1): 102- 111. DOI: 10.1136/gutjnl-2012-302779. |
| [12] |
KLINE A, WANG HY, LI YK, et al. Multimodal machine learning in precision health: A scoping review[J]. NPJ Digit Med, 2022, 5( 1): 171. DOI: 10.1038/s41746-022-00712-8. |
| [13] |
LIPKOVA J, CHEN RJ, CHEN BW, et al. Artificial intelligence for multimodal data integration in oncology[J]. Cancer Cell, 2022, 40( 10): 1095- 1110. DOI: 10.1016/j.ccell.2022.09.012. |
| [14] |
MOHSEN F, ALI H, HAJJ N EL, et al. Artificial intelligence-based methods for fusion of electronic health records and imaging data[J]. Sci Rep, 2022, 12( 1): 17981. DOI: 10.1038/s41598-022-22514-4. |
| [15] |
SHAO J, MA JC, ZHANG Q, et al. Predicting gene mutation status via artificial intelligence technologies based on multimodal integration(MMI) to advance precision oncology[J]. Semin Cancer Biol, 2023, 91: 1- 15. DOI: 10.1016/j.semcancer.2023.02.006. |
| [16] |
HUANG SC, PAREEK A, SEYYEDI S, et al. Fusion of medical imaging and electronic health records using deep learning: A systematic review and implementation guidelines[J]. NPJ Digit Med, 2020, 3: 136. DOI: 10.1038/s41746-020-00341-z. |
| [17] |
GLIEM N, AMMER-HERRMENAU C, ELLENRIEDER V, et al. Management of severe acute pancreatitis: An update[J]. Digestion, 2021, 102( 4): 503- 507. DOI: 10.1159/000506830. |
| [18] |
GAO X, LIN JX, WU AR, et al. Application of machine learning model based on XGBoost algorithm in early prediction of patients with acute severe pancreatitis[J]. Chin Crit Care Med, 2023, 35( 4): 421- 426. DOI: 10.3760/cma.j.cn121430-20221019-00930. |
| [19] |
CHEN ZY, WANG Y, ZHANG HL, et al. Deep learning models for severity prediction of acute pancreatitis in the early phase from abdominal nonenhanced computed tomography images[J]. Pancreas, 2023, 52( 1): e45- e53. DOI: 10.1097/MPA.0000000000002216. |
| [20] |
YIN MY, LIN JX, WANG Y, et al. Development and validation of a multimodal model in predicting severe acute pancreatitis based on radiomics and deep learning[J]. Int J Med Inform, 2024, 184: 105341. DOI: 10.1016/j.ijmedinf.2024.105341. |
| [21] |
KAMNITSAS K, LEDIG C, NEWCOMBE VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation[J]. Med Image Anal, 2017, 36: 61- 78. DOI: 10.1016/j.media.2016.10.004. |
| [22] |
LITJENS G, KOOI T, BEJNORDI BE, et al. A survey on deep learning in medical image analysis[J]. Med Image Anal, 2017, 42: 60- 88. DOI: 10.1016/j.media.2017.07.005. |



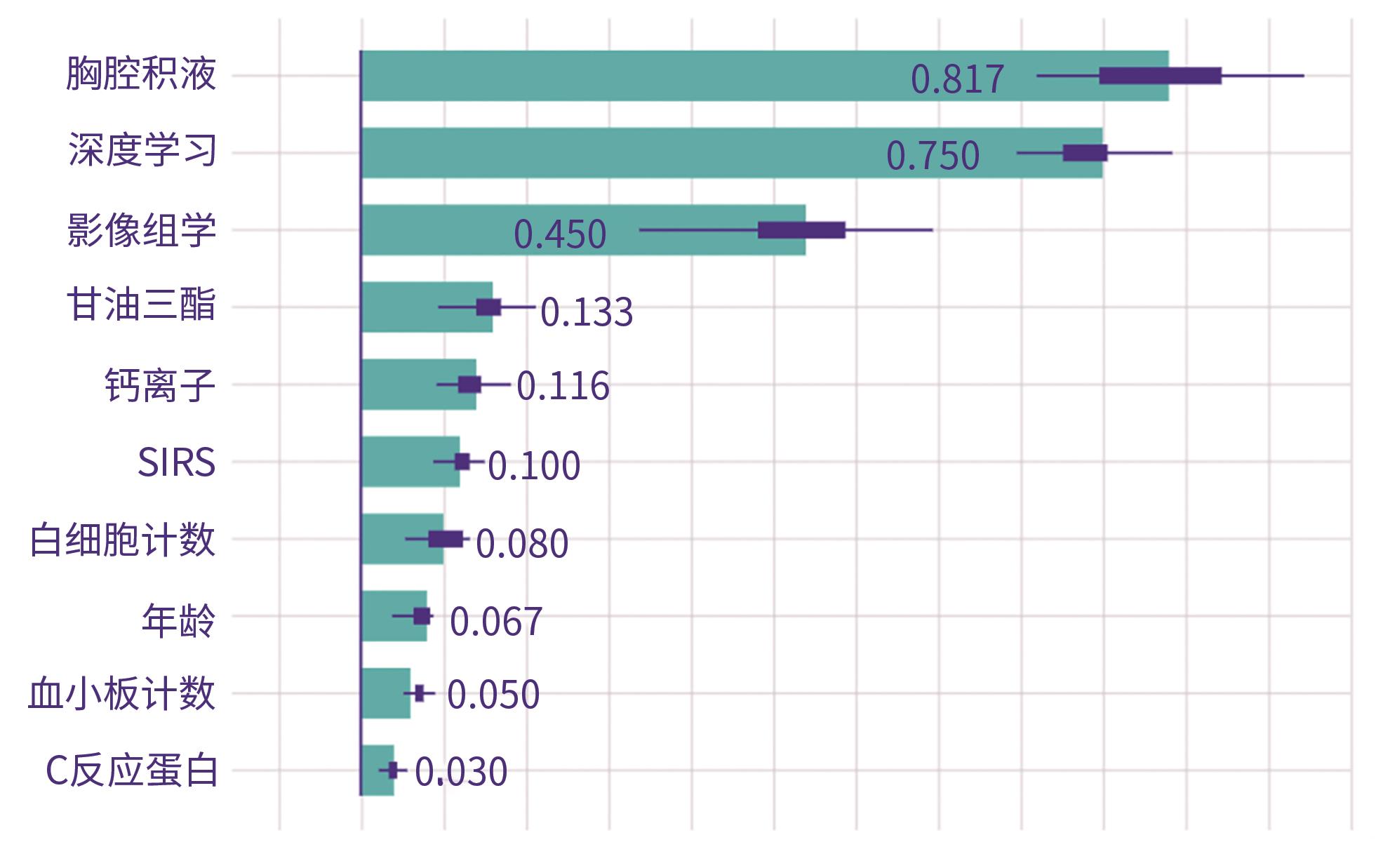




 DownLoad:
DownLoad: